

The eukaryotic genome presents a complex landscape where specific regions exhibit characteristics that promote gene duplications, which are pivotal for generating new genes and genetic diversity. Identifying these duplication hotspots and understanding the features of their surrounding genomic areas is essential for unraveling the diversity of molecular evolutionary mechanisms across taxa. My research focuses on developing tools and employing bioinformatic and phylogenetic approaches to address this challenge, with a particular interest in investigating parasitic protists.
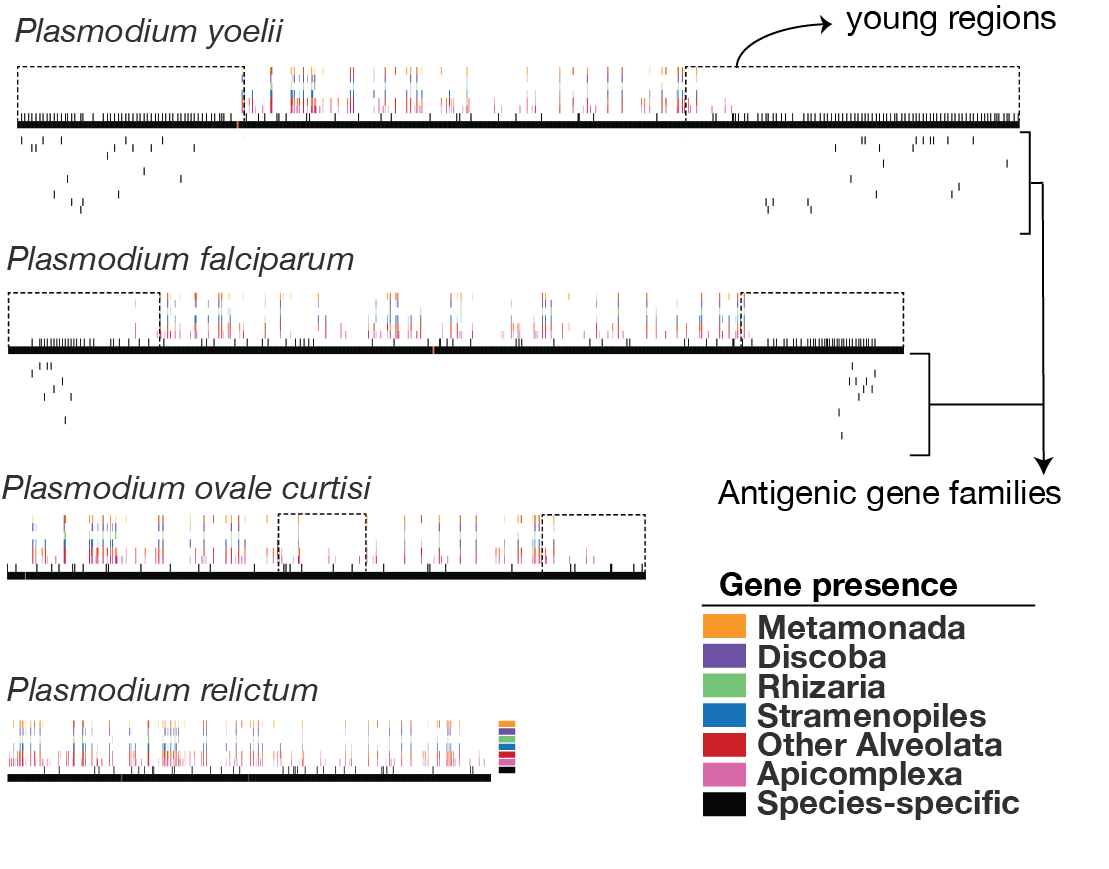
Parasitic protists have evolved a diverse array of molecular mechanisms aimed at increasing the diversity of antigenic gene families and evading host immune responses. I am particularly interested in studying these mechanisms and their impact on genomic evolution. The figure above illustrates exemplary maps of gene conservation across the chromosomes of various Plasmodium species, generated using the PhyloChromoMap tool (Cerón-Romero et al. 2018). In the figure, the horizontal black bars represent chromosomes, the vertical bars above the chromosomes depict gene conservation as heatmaps, and the vertical lines below the chromosomes indicate antigenic genes. These maps reveal varying degrees of evidence regarding the presence of mechanisms that induce antigenic variation through ectopic recombination of subtelomeres or chromosome ends. For instance, P. yoelii exhibits more evidence of this mechanism, as evidenced by the extended young regions in the subtelomeres containing numerous antigenic genes (Martínez-Eraso, et al. 2024).
Department of Biology
Case Western Reserve University
Clapp Hall Room 307
2080 Adelbert Road
Cleveland, OH 44106-7078
mario.ceronromero [at] case [dot] edu
© 2023. Mario A. Cerón-Romero